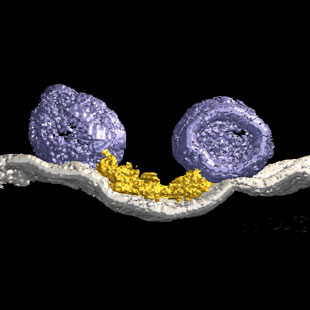
3-Dimensional model from the active zone of the frog's neuromuscular junction.
Reference:
Harlow, M. et al, (2001). The
architecture of active zone material
at the frog's neuromuscular junction.
Nature 409:479-484, 2001.
EM3D: Software for Electron Microscope Tomography
EM3D is an integrated software application designed to facilitate the analysis and visualization of electron microscope (EM) tomography data by cellular and molecular biologists. Such data are collected as a tilt series, a sequence of 2D electron micrographs taken at many tilt angles with respect to the electron beam.
Development of EM3D began in 1997 by Dr. Davis Ress who had joined the McMahan Lab to design, develop, and create an integrated software package that would take raw electron microscope image data to transform into individual surface models. Different from volume rendered models, individual objects could be viewed and analyzed individually or with other objects within the data set. Also, his objecive was to incorporate all the data manipulations remain within EM3D and not ported from one application to another. Most import for the general user, the software would run on most commonly used compter platforms. To facilitate this software development (dubbed EM3D as a working title) he used IDL, an interface descriptive language software, that could quickly develop tools and graphical user interfaces that were intuitive to an average user as well as logical in its work flow. By the time he left Stanford, EM3D was released as a fully functional application using IDL scripting.
EM3D features an integrated graphical user interface that automates most of the initial alignment and reconstruction of the tilt-series data to form a 3D volume. EM3D can also combine dual axis data sets, ie two data sets taken orthogonal to one another. These functions seamlessly connect with segmentation and model-generation tools that permit the user to extract easily and reliably specific structural components from the reconstructed volume. These segmentations are based on the stain density of the sample allowing the user to make objective decisions on the segmentation. The 3D models can then be viewed and analyzed using an extensive package of surface and volume rendering techniques.
EM3D also provides a set of analysis tools to quantify structural information from the models, including their moments, proximity relationships, and spatial reliability. Altogether, EM3D facilitates the analysis of 3D cell structure at the full resolution of a reconstructed volume (2-3 nm).
EM3D is available free-of-charge for most computer platforms (Mac OS 10.6, and Windows XP, Vista). This site provides software distribution and updates as well as tutorials, sample results, etc.
EM3D is being developed in the laboratory of Dr. U. J. McMahan, Professor of Biology at Texas A&M University (previously at Stanford University School of Medicine, Dept. of Neurobiology). EM3D is the joint effort of cellular and molecular cell biologists, computational biologists and engineers who use it daily.
U. J. McMahan |
|
Last modified:
3/1/22
This Human Brain Project/Neuroinformatics research
is funded by the
National Institute of Mental Health